Appendices
A: Other analyses & visualizations
Here is code for other representation of taxa abundance. These are raw R images that have not been gussied up.
We will create two different representations of relative abundance for each sample (arranged by host species) by major Classes—facet grid box-and-whisker plots and bar charts. We will generate each separately (and save) using an earlier relative abundance phyloseq object and then display the combined output.
Code for box-and-whisker plot
library(phyloseq)
mdata_phy_all <- tax_glom(ps_slv_filt_AVG, taxrank = "Class", NArm = FALSE)
# You can choose any taxonomic level here
mdata_phyrel_all <- transform_sample_counts(
mdata_phy_all, function(x) x / sum(x)
)
meltd_all <- psmelt(mdata_phyrel_all)
meltd_all$Class <- as.character(meltd_all$Class)
means <- ddply(meltd_all, ~Class, function(x) c(mean = mean(x$Abundance)))
# decending order
taxa_means <- means[order(-means$mean), ]
# ditch the sci notation
taxa_means <- format(taxa_means, scientific = FALSE)
# Here we conglomerate at 2%.
Other <- means[means$mean <= 0.026, ]$Class
meltd_all[meltd_all$Class %in% Other, ]$Class <- "Other"
samp_names <- aggregate(meltd_all$Abundance,
by = list(meltd_all$Sample), FUN = sum)[, 1]
.e <- environment()
meltd_all[, "Class"] <- factor(meltd_all[, "Class"],
sort(unique(meltd_all[, "Class"])))
meltd_all <- meltd_all[order(meltd_all[, "Class"]), ]
levels(meltd_all$Class)
# Here we order Classes by the Phylum they belong to.
meltd_all$Class <- factor(meltd_all$Class,
levels = c(
"Bacteroidia", "Clostridia", "Erysipelotrichia",
"Fusobacteriia", "Alphaproteobacteria",
"Deltaproteobacteria", "Gammaproteobacteria",
"Planctomycetacia", "Oxyphotobacteria", "Other"))
sup_fig1 <- qplot(data = meltd_all, x = Sp, y = Abundance, fill = Class,
geom = "boxplot", ylab = "Relative Abundance") +
theme(legend.position = "bottom") +
facet_grid(Class ~ ., scales = "free_y", space = "free_y") +
geom_jitter(width = 0.05) +
geom_point(colour = "black", fill = "white")
#+ guides(guide_legend(reverse = FALSE) )
sup_fig1 <- sup_fig1 +
scale_fill_manual(values = friend_pal) +
labs(x = "Host species", y = "Relative abundance (% total reads)")
pdf("DATA/PHYLOSEQ/FIGURES/OUTPUT/box_and_whisker.pdf")
sup_fig1
invisible(dev.off())Code for bar plot
Figure S1
sup_fig2 <- ggplot(meltd_all, aes_string(x = "Sample", y = "Abundance",
fill = "Class"), environment = .e, Ordered = TRUE)
sup_fig2 <- sup_fig2 + geom_bar(stat = "identity", position = "stack") +
facet_grid(Class ~ Sp, scales = "free", space = "free")
sup_fig2 <- sup_fig2 + scale_fill_manual(values = friend_pal)
# sup_fig2 <- sup_fig2 + theme(axis.text.x = element_text(angle = -90,
# hjust = 0))
sup_fig2 <- sup_fig2 + theme(axis.text.x = element_blank())
sup_fig2 <- sup_fig2 +
guides(fill = guide_legend(override.aes = list(colour = NULL),
reverse = FALSE)) +
theme(legend.key = element_rect(colour = "black"),
legend.position = "bottom") +
labs(x = "Individual samples", y = "Relative abundance (% total reads)")
pdf("DATA/PHYLOSEQ/FIGURES/OUTPUT/Figure_S1.pdf")
sup_fig2
invisible(dev.off())grid.arrange(sup_fig1, sup_fig2, ncol = 2)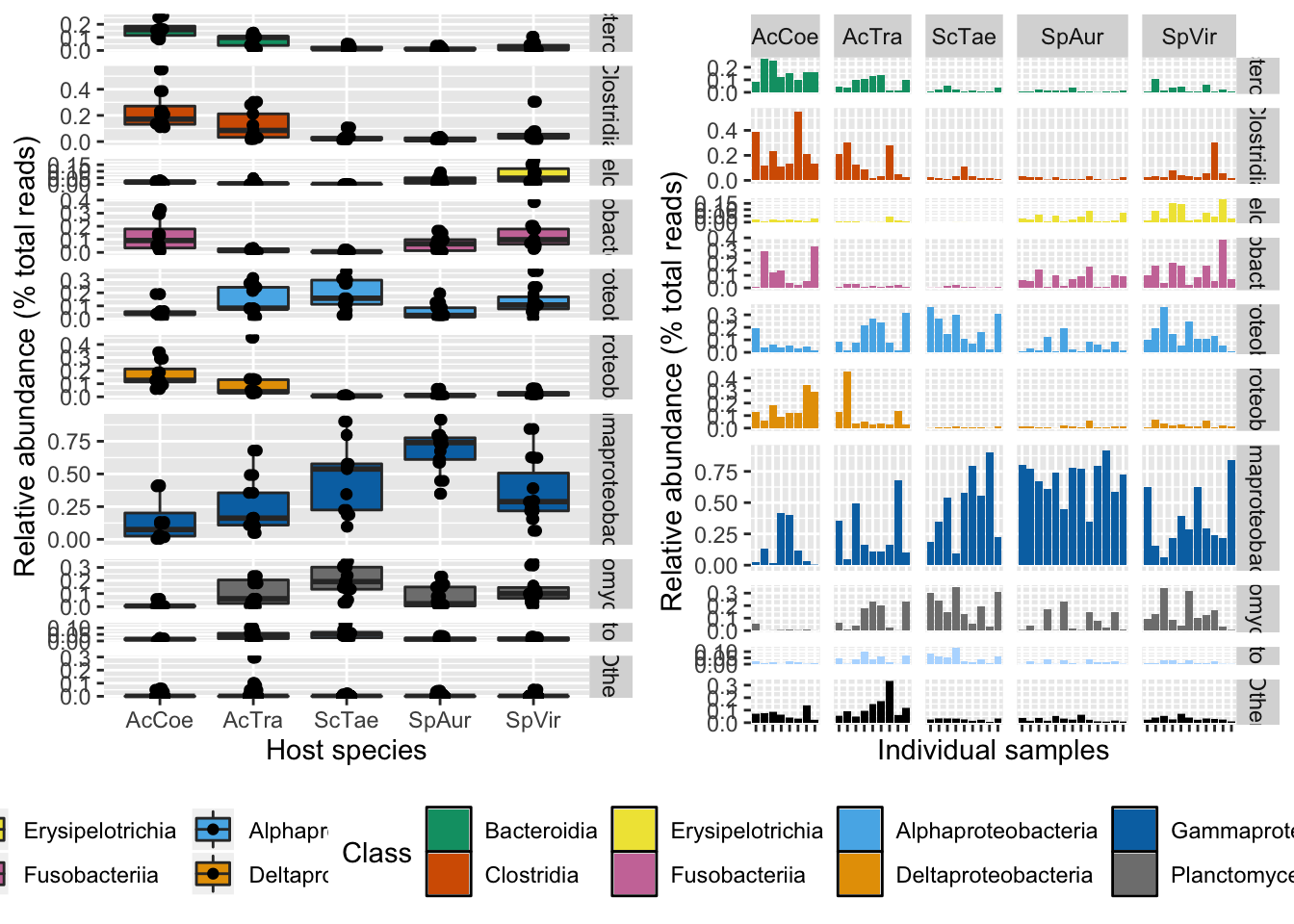
Figure S1. Relative abundance of major Classes by sample
B: Tools & resources used in this workflow
Specific tools
- phyloseq as the primary analytical package.
- LEfSe to identify differentially abundant (DA) amplicon sequence variants (ASV) across host fish species.
- MicrobiomeAnalyst to conduct LEfSe analysis.
- BLASTn and Silva ACT to identify closest hits to DA ASVs.
- RAxML for phylogenetic inference of DA ASVs and closest hits.
- iTOL for visualization of tree and associated metadata.
Other valuable resources
- R Markdown: The Definitive Guide
- knitr tutorials. Fantastic site!
- Microbiota analysis in R, UCR Workshop 2018 nicely documented workflow with examples.
C: Submitting sequencing data to public archives
It is now time to submit the data to your favorite sequence read archive. We submitted out data to the European Nucleotide Archive (ENA). The ENA does not like RAW data and prefers to have primers removed. So we submitted the trimmed Fastq files to the ENA. You can find these data under the study accession number PRJEB28397. The raw, raw files are available on the project’s figshare site.
To submit to the ENA you need two data tables (plus your sequence data). The first file describes the samples and the second file describes the sequencing data. The original files can be found on the figshare site with the raw data.
D: Specific R package & versions
Below are the specific packages and versions used in this workflow using both sessionInfo() and devtools::session_info().
Show/hide HYDRA TRIMMOMATIC job script
```r
sessionInfo()
```
```
## R version 3.6.1 (2019-07-05)
## Platform: x86_64-apple-darwin15.6.0 (64-bit)
## Running under: macOS Catalina 10.15.3
##
## Matrix products: default
## BLAS: /System/Library/Frameworks/Accelerate.framework/Versions/A/Frameworks/vecLib.framework/Versions/A/libBLAS.dylib
## LAPACK: /Library/Frameworks/R.framework/Versions/3.6/Resources/lib/libRlapack.dylib
##
## Random number generation:
## RNG: Mersenne-Twister
## Normal: Inversion
## Sample: Rounding
##
## locale:
## [1] en_US.UTF-8/en_US.UTF-8/en_US.UTF-8/C/en_US.UTF-8/en_US.UTF-8
##
## attached base packages:
## [1] grid stats4 parallel stats graphics
## [6] grDevices utils datasets methods base
##
## other attached packages:
## [1] whoppeR_0.0.0.9022
## [2] rsvg_1.3
## [3] gdtools_0.2.1
## [4] svglite_1.2.3
## [5] leaflet_2.0.3.9000
## [6] gapminder_0.3.0
## [7] phyloseq_1.28.0
## [8] treeio_1.8.2
## [9] tidytree_0.3.1
## [10] ggtree_1.16.6
## [11] picante_1.8.1
## [12] nlme_3.1-144
## [13] gdata_2.18.0
## [14] heatmap3_1.1.6
## [15] circlize_0.4.8
## [16] ComplexHeatmap_2.1.0
## [17] ggpubr_0.2.5
## [18] magrittr_1.5
## [19] ggthemes_4.2.0
## [20] seqinr_3.6-1
## [21] ips_0.0.11
## [22] ape_5.3
## [23] TeachingDemos_2.10
## [24] pairwiseAdonis_0.0.1
## [25] cluster_2.1.0
## [26] plotly_4.9.2
## [27] RCurl_1.98-1.1
## [28] bitops_1.0-6
## [29] svgPanZoom_0.3.4
## [30] gridExtra_2.3
## [31] forcats_0.4.0
## [32] stringr_1.4.0
## [33] dplyr_0.8.4
## [34] purrr_0.3.3
## [35] readr_1.3.1
## [36] tidyr_1.0.2
## [37] tibble_2.1.3
## [38] tidyverse_1.3.0
## [39] formatR_1.7
## [40] pander_0.6.3
## [41] rmarkdown_2.1
## [42] DT_0.12
## [43] data.table_1.12.8
## [44] kableExtra_1.1.0
## [45] knitr_1.28
## [46] rstudioapi_0.11
## [47] reshape2_1.4.3
## [48] scales_1.1.0
## [49] vegan_2.5-6
## [50] lattice_0.20-38
## [51] permute_0.9-5
## [52] plyr_1.8.5
## [53] ggplot2_3.2.1
## [54] ShortRead_1.42.0
## [55] GenomicAlignments_1.20.1
## [56] SummarizedExperiment_1.14.1
## [57] DelayedArray_0.10.0
## [58] matrixStats_0.55.0
## [59] Biobase_2.44.0
## [60] Rsamtools_2.0.3
## [61] GenomicRanges_1.36.1
## [62] GenomeInfoDb_1.20.0
## [63] Biostrings_2.52.0
## [64] XVector_0.24.0
## [65] IRanges_2.18.3
## [66] S4Vectors_0.22.1
## [67] BiocParallel_1.18.1
## [68] BiocGenerics_0.30.0
## [69] dada2_1.12.1
## [70] Rcpp_1.0.3
##
## loaded via a namespace (and not attached):
## [1] tidyselect_1.0.0 htmlwidgets_1.5.1
## [3] devtools_2.2.1 munsell_0.5.0
## [5] codetools_0.2-16 withr_2.1.2
## [7] colorspace_1.4-1 ggsignif_0.6.0
## [9] GenomeInfoDbData_1.2.1 hwriter_1.3.2
## [11] rhdf5_2.28.1 rprojroot_1.3-2
## [13] vctrs_0.2.2 generics_0.0.2
## [15] xfun_0.12 fastcluster_1.1.25
## [17] R6_2.4.1 clue_0.3-57
## [19] assertthat_0.2.1 promises_1.1.0
## [21] gtable_0.3.0 processx_3.4.2
## [23] phangorn_2.5.5 rlang_0.4.4
## [25] systemfonts_0.1.1 GlobalOptions_0.1.1
## [27] splines_3.6.1 lazyeval_0.2.2
## [29] broom_0.5.4 BiocManager_1.30.10
## [31] yaml_2.2.1 modelr_0.1.5
## [33] crosstalk_1.0.0 backports_1.1.5
## [35] httpuv_1.5.2 tools_3.6.1
## [37] usethis_1.5.1 ellipsis_0.3.0
## [39] biomformat_1.12.0 RColorBrewer_1.1-2
## [41] sessioninfo_1.1.1 zlibbioc_1.30.0
## [43] ps_1.3.2 prettyunits_1.1.1
## [45] GetoptLong_0.1.8 haven_2.2.0
## [47] fs_1.3.1 reprex_0.3.0
## [49] pkgload_1.0.2 xtable_1.8-4
## [51] mime_0.9 hms_0.5.3
## [53] evaluate_0.14 XML_3.99-0.3
## [55] jpeg_0.1-8.1 readxl_1.3.1
## [57] shape_1.4.4 testthat_2.3.1
## [59] compiler_3.6.1 crayon_1.3.4
## [61] htmltools_0.4.0 later_1.0.0
## [63] mgcv_1.8-31 RcppParallel_4.4.4
## [65] lubridate_1.7.4 DBI_1.1.0
## [67] dbplyr_1.4.2 MASS_7.3-51.5
## [69] Matrix_1.2-18 ade4_1.7-15
## [71] cli_2.0.1 quadprog_1.5-8
## [73] igraph_1.2.4.2 pkgconfig_2.0.3
## [75] rvcheck_0.1.7 xml2_1.2.2
## [77] foreach_1.4.8 multtest_2.40.0
## [79] webshot_0.5.2 rvest_0.3.5
## [81] callr_3.4.2 digest_0.6.24
## [83] cellranger_1.1.0 fastmatch_1.1-0
## [85] shiny_1.4.0 gtools_3.8.1
## [87] rjson_0.2.20 lifecycle_0.1.0
## [89] jsonlite_1.6.1 Rhdf5lib_1.6.3
## [91] desc_1.2.0 viridisLite_0.3.0
## [93] fansi_0.4.1 pillar_1.4.3
## [95] fastmap_1.0.1 httr_1.4.1
## [97] pkgbuild_1.0.6 survival_3.1-8
## [99] glue_1.3.1 remotes_2.1.1
## [101] png_0.1-7 iterators_1.0.12
## [103] stringi_1.4.6 latticeExtra_0.6-29
## [105] memoise_1.1.0
```
```r
devtools::session_info()
```
```
## ─ Session info ──────────────────────────────────────────
## setting value
## version R version 3.6.1 (2019-07-05)
## os macOS Catalina 10.15.3
## system x86_64, darwin15.6.0
## ui RStudio
## language (EN)
## collate en_US.UTF-8
## ctype en_US.UTF-8
## tz America/Panama
## date 2020-03-17
##
## ─ Packages ──────────────────────────────────────────────
## package * version date lib
## ade4 1.7-15 2020-02-13 [1]
## ape * 5.3 2019-03-17 [1]
## assertthat 0.2.1 2019-03-21 [1]
## backports 1.1.5 2019-10-02 [1]
## Biobase * 2.44.0 2019-05-02 [1]
## BiocGenerics * 0.30.0 2019-05-02 [1]
## BiocManager 1.30.10 2019-11-16 [1]
## BiocParallel * 1.18.1 2019-08-06 [1]
## biomformat 1.12.0 2019-05-02 [1]
## Biostrings * 2.52.0 2019-05-02 [1]
## bitops * 1.0-6 2013-08-17 [1]
## broom 0.5.4 2020-01-27 [1]
## callr 3.4.2 2020-02-12 [1]
## cellranger 1.1.0 2016-07-27 [1]
## circlize * 0.4.8 2019-09-08 [1]
## cli 2.0.1 2020-01-08 [1]
## clue 0.3-57 2019-02-25 [1]
## cluster * 2.1.0 2019-06-19 [1]
## codetools 0.2-16 2018-12-24 [1]
## colorspace 1.4-1 2019-03-18 [1]
## ComplexHeatmap * 2.1.0 2019-09-10 [1]
## crayon 1.3.4 2017-09-16 [1]
## crosstalk 1.0.0 2016-12-21 [1]
## dada2 * 1.12.1 2019-05-14 [1]
## data.table * 1.12.8 2019-12-09 [1]
## DBI 1.1.0 2019-12-15 [1]
## dbplyr 1.4.2 2019-06-17 [1]
## DelayedArray * 0.10.0 2019-05-02 [1]
## desc 1.2.0 2018-05-01 [1]
## devtools 2.2.1 2019-09-24 [1]
## digest 0.6.24 2020-02-12 [1]
## dplyr * 0.8.4 2020-01-31 [1]
## DT * 0.12 2020-02-05 [1]
## ellipsis 0.3.0 2019-09-20 [1]
## evaluate 0.14 2019-05-28 [1]
## fansi 0.4.1 2020-01-08 [1]
## fastcluster 1.1.25 2018-06-07 [1]
## fastmap 1.0.1 2019-10-08 [1]
## fastmatch 1.1-0 2017-01-28 [1]
## forcats * 0.4.0 2019-02-17 [1]
## foreach 1.4.8 2020-02-09 [1]
## formatR * 1.7 2019-06-11 [1]
## fs 1.3.1 2019-05-06 [1]
## gapminder * 0.3.0 2017-10-31 [1]
## gdata * 2.18.0 2017-06-06 [1]
## gdtools * 0.2.1 2019-10-14 [1]
## generics 0.0.2 2018-11-29 [1]
## GenomeInfoDb * 1.20.0 2019-05-02 [1]
## GenomeInfoDbData 1.2.1 2019-09-10 [1]
## GenomicAlignments * 1.20.1 2019-06-18 [1]
## GenomicRanges * 1.36.1 2019-09-06 [1]
## GetoptLong 0.1.8 2020-01-08 [1]
## ggplot2 * 3.2.1 2019-08-10 [1]
## ggpubr * 0.2.5 2020-02-13 [1]
## ggsignif 0.6.0 2019-08-08 [1]
## ggthemes * 4.2.0 2019-05-13 [1]
## ggtree * 1.16.6 2019-08-26 [1]
## GlobalOptions 0.1.1 2019-09-30 [1]
## glue 1.3.1 2019-03-12 [1]
## gridExtra * 2.3 2017-09-09 [1]
## gtable 0.3.0 2019-03-25 [1]
## gtools 3.8.1 2018-06-26 [1]
## haven 2.2.0 2019-11-08 [1]
## heatmap3 * 1.1.6 2019-03-22 [1]
## hms 0.5.3 2020-01-08 [1]
## htmltools 0.4.0 2019-10-04 [1]
## htmlwidgets 1.5.1 2019-10-08 [1]
## httpuv 1.5.2 2019-09-11 [1]
## httr 1.4.1 2019-08-05 [1]
## hwriter 1.3.2 2014-09-10 [1]
## igraph 1.2.4.2 2019-11-27 [1]
## ips * 0.0.11 2019-07-04 [1]
## IRanges * 2.18.3 2019-09-24 [1]
## iterators 1.0.12 2019-07-26 [1]
## jpeg 0.1-8.1 2019-10-24 [1]
## jsonlite 1.6.1 2020-02-02 [1]
## kableExtra * 1.1.0 2019-03-16 [1]
## knitr * 1.28 2020-02-06 [1]
## later 1.0.0 2019-10-04 [1]
## lattice * 0.20-38 2018-11-04 [1]
## latticeExtra 0.6-29 2019-12-19 [1]
## lazyeval 0.2.2 2019-03-15 [1]
## leaflet * 2.0.3.9000 2019-12-11 [1]
## lifecycle 0.1.0 2019-08-01 [1]
## lubridate 1.7.4 2018-04-11 [1]
## magrittr * 1.5 2014-11-22 [1]
## MASS 7.3-51.5 2019-12-20 [1]
## Matrix 1.2-18 2019-11-27 [1]
## matrixStats * 0.55.0 2019-09-07 [1]
## memoise 1.1.0 2017-04-21 [1]
## mgcv 1.8-31 2019-11-09 [1]
## mime 0.9 2020-02-04 [1]
## modelr 0.1.5 2019-08-08 [1]
## multtest 2.40.0 2019-05-02 [1]
## munsell 0.5.0 2018-06-12 [1]
## nlme * 3.1-144 2020-02-06 [1]
## pairwiseAdonis * 0.0.1 2019-09-10 [1]
## pander * 0.6.3 2018-11-06 [1]
## permute * 0.9-5 2019-03-12 [1]
## phangorn 2.5.5 2019-06-19 [1]
## phyloseq * 1.28.0 2019-05-02 [1]
## picante * 1.8.1 2020-02-13 [1]
## pillar 1.4.3 2019-12-20 [1]
## pkgbuild 1.0.6 2019-10-09 [1]
## pkgconfig 2.0.3 2019-09-22 [1]
## pkgload 1.0.2 2018-10-29 [1]
## plotly * 4.9.2 2020-02-12 [1]
## plyr * 1.8.5 2019-12-10 [1]
## png 0.1-7 2013-12-03 [1]
## prettyunits 1.1.1 2020-01-24 [1]
## processx 3.4.2 2020-02-09 [1]
## promises 1.1.0 2019-10-04 [1]
## ps 1.3.2 2020-02-13 [1]
## purrr * 0.3.3 2019-10-18 [1]
## quadprog 1.5-8 2019-11-20 [1]
## R6 2.4.1 2019-11-12 [1]
## RColorBrewer 1.1-2 2014-12-07 [1]
## Rcpp * 1.0.3 2019-11-08 [1]
## RcppParallel 4.4.4 2019-09-27 [1]
## RCurl * 1.98-1.1 2020-01-19 [1]
## readr * 1.3.1 2018-12-21 [1]
## readxl 1.3.1 2019-03-13 [1]
## remotes 2.1.1 2020-02-15 [1]
## reprex 0.3.0 2019-05-16 [1]
## reshape2 * 1.4.3 2017-12-11 [1]
## rhdf5 2.28.1 2019-10-10 [1]
## Rhdf5lib 1.6.3 2019-10-15 [1]
## rjson 0.2.20 2018-06-08 [1]
## rlang 0.4.4 2020-01-28 [1]
## rmarkdown * 2.1 2020-01-20 [1]
## rprojroot 1.3-2 2018-01-03 [1]
## Rsamtools * 2.0.3 2019-10-10 [1]
## rstudioapi * 0.11 2020-02-07 [1]
## rsvg * 1.3 2018-05-10 [1]
## rvcheck 0.1.7 2019-11-29 [1]
## rvest 0.3.5 2019-11-08 [1]
## S4Vectors * 0.22.1 2019-09-09 [1]
## scales * 1.1.0 2019-11-18 [1]
## seqinr * 3.6-1 2019-09-07 [1]
## sessioninfo 1.1.1 2018-11-05 [1]
## shape 1.4.4 2018-02-07 [1]
## shiny 1.4.0 2019-10-10 [1]
## ShortRead * 1.42.0 2019-05-02 [1]
## stringi 1.4.6 2020-02-17 [1]
## stringr * 1.4.0 2019-02-10 [1]
## SummarizedExperiment * 1.14.1 2019-07-31 [1]
## survival 3.1-8 2019-12-03 [1]
## svglite * 1.2.3 2020-02-07 [1]
## svgPanZoom * 0.3.4 2020-02-15 [1]
## systemfonts 0.1.1 2019-07-01 [1]
## TeachingDemos * 2.10 2016-02-12 [1]
## testthat 2.3.1 2019-12-01 [1]
## tibble * 2.1.3 2019-06-06 [1]
## tidyr * 1.0.2 2020-01-24 [1]
## tidyselect 1.0.0 2020-01-27 [1]
## tidytree * 0.3.1 2019-12-14 [1]
## tidyverse * 1.3.0 2019-11-21 [1]
## treeio * 1.8.2 2019-08-21 [1]
## usethis 1.5.1 2019-07-04 [1]
## vctrs 0.2.2 2020-01-24 [1]
## vegan * 2.5-6 2019-09-01 [1]
## viridisLite 0.3.0 2018-02-01 [1]
## webshot 0.5.2 2019-11-22 [1]
## whoppeR * 0.0.0.9022 2019-11-26 [1]
## withr 2.1.2 2018-03-15 [1]
## xfun 0.12 2020-01-13 [1]
## XML 3.99-0.3 2020-01-20 [1]
## xml2 1.2.2 2019-08-09 [1]
## xtable 1.8-4 2019-04-21 [1]
## XVector * 0.24.0 2019-05-02 [1]
## yaml 2.2.1 2020-02-01 [1]
## zlibbioc 1.30.0 2019-05-02 [1]
## source
## CRAN (R 3.6.0)
## CRAN (R 3.6.0)
## CRAN (R 3.6.0)
## CRAN (R 3.6.0)
## Bioconductor
## Bioconductor
## CRAN (R 3.6.0)
## Bioconductor
## Bioconductor
## Bioconductor
## CRAN (R 3.6.0)
## CRAN (R 3.6.0)
## CRAN (R 3.6.0)
## CRAN (R 3.6.0)
## CRAN (R 3.6.0)
## CRAN (R 3.6.0)
## CRAN (R 3.6.0)
## CRAN (R 3.6.1)
## CRAN (R 3.6.1)
## CRAN (R 3.6.0)
## Github (jokergoo/ComplexHeatmap@35d1d20)
## CRAN (R 3.6.0)
## CRAN (R 3.6.0)
## Bioconductor
## CRAN (R 3.6.0)
## CRAN (R 3.6.0)
## CRAN (R 3.6.0)
## Bioconductor
## CRAN (R 3.6.0)
## CRAN (R 3.6.0)
## CRAN (R 3.6.0)
## CRAN (R 3.6.0)
## CRAN (R 3.6.0)
## CRAN (R 3.6.0)
## CRAN (R 3.6.0)
## CRAN (R 3.6.0)
## CRAN (R 3.6.0)
## CRAN (R 3.6.0)
## CRAN (R 3.6.0)
## CRAN (R 3.6.0)
## CRAN (R 3.6.0)
## CRAN (R 3.6.0)
## CRAN (R 3.6.0)
## CRAN (R 3.6.0)
## CRAN (R 3.6.0)
## CRAN (R 3.6.0)
## CRAN (R 3.6.0)
## Bioconductor
## Bioconductor
## Bioconductor
## Bioconductor
## CRAN (R 3.6.0)
## CRAN (R 3.6.0)
## CRAN (R 3.6.0)
## CRAN (R 3.6.0)
## CRAN (R 3.6.0)
## Bioconductor
## CRAN (R 3.6.0)
## CRAN (R 3.6.0)
## CRAN (R 3.6.0)
## CRAN (R 3.6.0)
## CRAN (R 3.6.0)
## CRAN (R 3.6.0)
## CRAN (R 3.6.0)
## CRAN (R 3.6.0)
## CRAN (R 3.6.0)
## CRAN (R 3.6.0)
## CRAN (R 3.6.0)
## CRAN (R 3.6.0)
## CRAN (R 3.6.0)
## CRAN (R 3.6.0)
## CRAN (R 3.6.0)
## Bioconductor
## CRAN (R 3.6.0)
## CRAN (R 3.6.0)
## CRAN (R 3.6.0)
## CRAN (R 3.6.0)
## CRAN (R 3.6.0)
## CRAN (R 3.6.0)
## CRAN (R 3.6.1)
## CRAN (R 3.6.0)
## CRAN (R 3.6.0)
## Github (rstudio/leaflet@c19b0fb)
## CRAN (R 3.6.0)
## CRAN (R 3.6.0)
## CRAN (R 3.6.0)
## CRAN (R 3.6.0)
## CRAN (R 3.6.0)
## CRAN (R 3.6.0)
## CRAN (R 3.6.0)
## CRAN (R 3.6.0)
## CRAN (R 3.6.0)
## CRAN (R 3.6.0)
## Bioconductor
## CRAN (R 3.6.0)
## CRAN (R 3.6.0)
## Github (pmartinezarbizu/pairwiseAdonis@6e09713)
## CRAN (R 3.6.0)
## CRAN (R 3.6.0)
## CRAN (R 3.6.0)
## Bioconductor
## CRAN (R 3.6.0)
## CRAN (R 3.6.0)
## CRAN (R 3.6.0)
## CRAN (R 3.6.0)
## CRAN (R 3.6.0)
## CRAN (R 3.6.0)
## CRAN (R 3.6.0)
## CRAN (R 3.6.0)
## CRAN (R 3.6.0)
## CRAN (R 3.6.0)
## CRAN (R 3.6.0)
## CRAN (R 3.6.0)
## CRAN (R 3.6.0)
## CRAN (R 3.6.0)
## CRAN (R 3.6.0)
## CRAN (R 3.6.0)
## CRAN (R 3.6.0)
## CRAN (R 3.6.0)
## CRAN (R 3.6.0)
## CRAN (R 3.6.0)
## CRAN (R 3.6.0)
## CRAN (R 3.6.0)
## CRAN (R 3.6.0)
## CRAN (R 3.6.0)
## Bioconductor
## Bioconductor
## CRAN (R 3.6.0)
## CRAN (R 3.6.0)
## CRAN (R 3.6.0)
## CRAN (R 3.6.0)
## Bioconductor
## CRAN (R 3.6.0)
## CRAN (R 3.6.0)
## CRAN (R 3.6.0)
## CRAN (R 3.6.0)
## Bioconductor
## CRAN (R 3.6.0)
## CRAN (R 3.6.0)
## CRAN (R 3.6.0)
## CRAN (R 3.6.0)
## CRAN (R 3.6.0)
## Bioconductor
## CRAN (R 3.6.0)
## CRAN (R 3.6.0)
## Bioconductor
## CRAN (R 3.6.0)
## CRAN (R 3.6.0)
## CRAN (R 3.6.0)
## CRAN (R 3.6.0)
## CRAN (R 3.6.0)
## CRAN (R 3.6.0)
## CRAN (R 3.6.0)
## CRAN (R 3.6.0)
## CRAN (R 3.6.0)
## CRAN (R 3.6.0)
## CRAN (R 3.6.0)
## Bioconductor
## CRAN (R 3.6.0)
## CRAN (R 3.6.0)
## CRAN (R 3.6.0)
## CRAN (R 3.6.0)
## CRAN (R 3.6.0)
## Github (wjhopper/whoppeR@38319f6)
## CRAN (R 3.6.0)
## CRAN (R 3.6.0)
## CRAN (R 3.6.0)
## CRAN (R 3.6.0)
## CRAN (R 3.6.0)
## Bioconductor
## CRAN (R 3.6.0)
## Bioconductor
##
## [1] /Library/Frameworks/R.framework/Versions/3.6/Resources/library
```